DNA testing is on the rise, with companies like Ancestry DNA, My Heritage, and 23andme all making direct-to-consumer genetic testing more accessible. But there are a few different kinds of tests out on the market and they all offer different results. If you’re looking into a rare condition, whole genome sequencing might be necessary, but if you’re just looking into your ancestry or for general health insights, exome sequencing is likely to be all you’ll need. Read on to discover the differences between the tests and their uses.
What are the two main types of DNA testing?
There are two main types of DNA testing that are used in research and in direct-to-consumer tests: genotyping and sequencing.
Genotyping

Genotyping tests for a small number of positions in the DNA that are selected ahead of time. In humans, most genotyping arrays cover 500,000-1,000,000 positions, which are chosen because they are known to be variable between different people.
On the other hand, sequencing reads full pieces of DNA (rather than testing individual positions), so it results in more information, and does not require picking positions to test ahead of time. However, it is also much more expensive.
Within these two broad categories, there are many other different flavours of test. For example, there are hundreds of different genotype arrays – each array covers different positions in the genome and may be used for slightly different purposes. An example of this is the Illumina Global Screening array, which is very popular with direct-to-consumer genetic testing companies; this genotype array covers around 640,000 positions. Another example is the UKBiobank Array, which was used to test 500,000 participants in the UK biobank (as well as many other research projects) and covers 820,000 positions.
Different types of sequencing
Within the category of sequencing, there are three main tests that are used for research and diagnostic testing, as well as increasingly in direct-to-consumer testing.
Targeted gene panels
Targeted gene panels involve pre-selecting a set of sequences (usually protein-coding genes) to test in order to look for known disease-causing genetic variants. Targeted Gene Panels are a two step process where DNA is first filtered to capture only the pre-selected set of sequences or "targets." Then the captured DNA is sent off to be sequenced.
For example, a person with a family history of breast cancer may be tested for a panel of a few dozen genes that are known to cause breast cancer. As another example, a child or young adult who has unexplained epilepsy/seizures would be tested for a completely different set of genes known to cause epilepsy. There are hundreds of genes that are known to cause epilepsy when their sequence is changed, and an epilepsy gene panel would test of all of these genes at once.
For diagnosing rare and chronic disease, gene panels can often find the genetic variant causing the condition, but sometimes result in a "diagnostic odyssey" where nothing is found in the panel, and further testing needs to be done, sometimes taking years to find the cause of disease. For this reason, panel sequencing has started to be replaced by exome sequencing for many genetic disorders.
Exome sequencing

Exome sequencing involves a two-step process, similar to panel sequencing, where the DNA "filtered" to capture only DNA that maps to one of the 20,000 protein-coding genes in the genome. The second step is to sequence the DNA that maps to a protein coding gene, which corresponds to ~2% of the total DNA.
Exome sequencing is more expensive than panel sequencing, but has the incredible advantage of helping many hard-to-diagnose patients get answers and potentially treatment, faster. This kind of sequencing is also useful in a research context for finding genetic variants that influence disease risk or affect common traits.
However, the main drawback of exome sequencing is that it misses 98% of the DNA. This can be the cause of missed diagnoses in rare or chronic disorders, when the mutation is not in the ~2% of the DNA that codes for proteins. Exome sequencing can also make it more challenging to detect deletions or duplications of DNA. For both of these reasons, many research projects and diagnostic sequencing efforts such as the 100,000 genomes project, are now turning to whole genome sequencing.
Whole genome sequencing
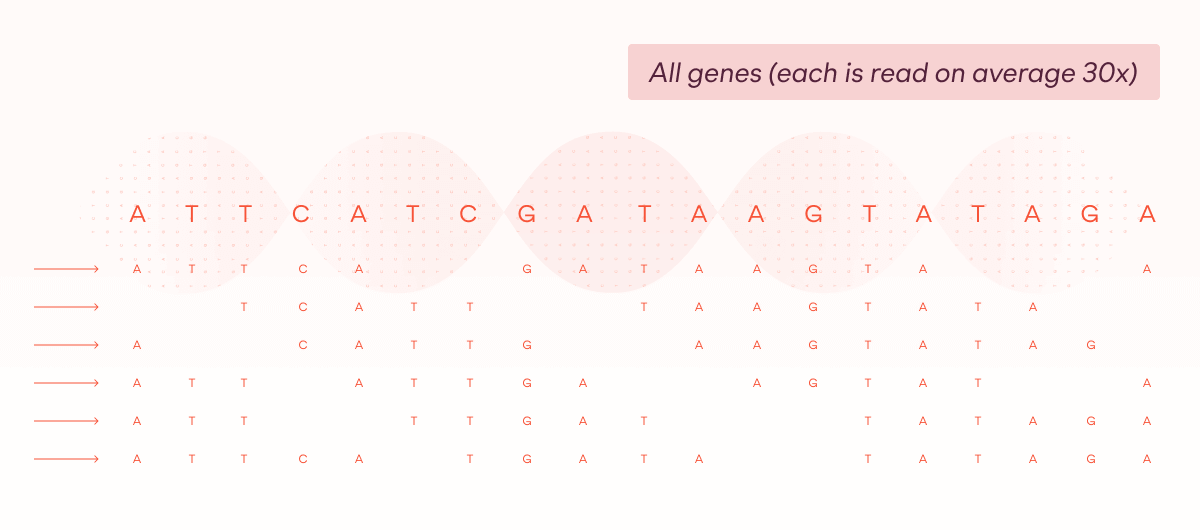
Whole genome sequencing does not require any filtering step – instead, the DNA is sent straight for sequencing, and there are on average approximately 30 reads of every position of the DNA. This gives a really accurate representation of the DNA sequence for nearly all of the 6.4 billion bases.
The majority of whole genome sequencing today uses "short read" technology – this means that about 100-150 bases of DNA are read at a time. As these reads overlap each other, they can be stitched together to form a nearly complete picture.
Other approaches
Besides the approaches covered above, there are others you may have heard of, specifically "low pass" whole genome sequencing and long read sequencing:
Low pass sequencing is a form of whole genome sequencing, but on average every base is covered by less than one sequencing read. In theory, statistical tools can be applied to piece together the missing sequence, making the data comparable to genotyping arrays, but with no need to pre-select positions to test. However, it is in no way a replacement for whole genome sequencing or exome sequencing.
Long read sequencing is a very exciting form of whole genome sequencing that reads very long stretches of DNA – sometimes tens of thousands of bases at a time, in contrast with the more commonly used short read sequencing. One big advantage of long read sequencing is that it can help to better detect deletions and duplications in DNA, which can be the cause of severe genetic conditions, as well as more subtle variations in traits or disease risk.
Another advantage of long read sequencing is that some technologies get information about methylation (the modifications to the DNA bases, sometimes referred to as ‘epigenetics’) at the same time as the DNA. Long read whole genome sequencing is still more expensive than short read sequencing, but the cost of long read sequencing is likely to continue to drop, and more uses of the technology will emerge.

